Lipids, lipid classes, lipid-Omics and the lipid-Ome:
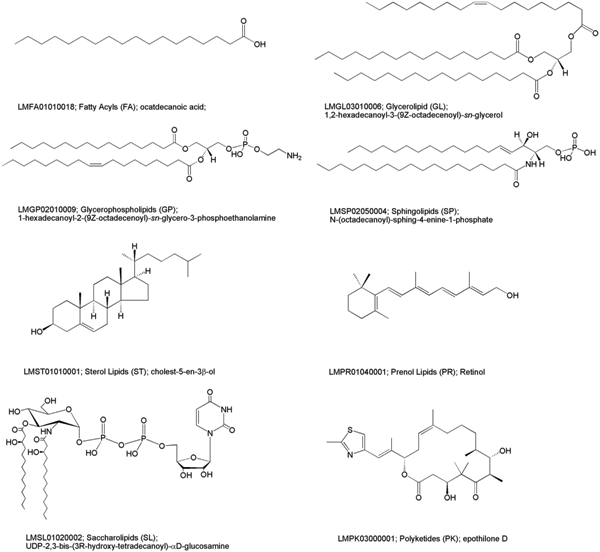
Lipids are a distinct and diverse set of small molecules consisting of eight general compound classes including (i) fatty acyls, (ii) glycerolipids, (ii) glycerophospholipids, (iv) sphingolipids, (v) sterol lipids, (vi) prenol lipids, (vii) saccharolipids and (viii) polyketides. For better classification 81 general lipid classes and 276 sub-categories were generated as a nomenclature by the LipidMaps consortium.
Picture (CC-by) thanks to: Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Murphy RC, Raetz CRH, Russell DW: LMSD: LIPID MAPS structure database. Nucleic Acids Research 2007, 35(1):D527-D532. ; doi:10.1093/nar/gkl838
Technologies:
In principle most of the analytical techniques can be used to analyze lipids including fatty acids, phospholipids, sphingolipids, triglycerides and steroids. Routinely thin layer chromatography (TLC) was used for such purposes and is now replaced by faster technologies with better resolution.
- GC-MS for low molecular weight lipids of all different classes (<500 Da) see our GC-TOF technology platform using BinBase and SetupX we can routinely profile around 80 lipid compounds (out of 800) in one single run and semi-quantify those lipid-like substances contained in the FiehnLib mass spectral and retention index database with high reliability. This equals a coverage of 11% of the compounds (based on a 500 Da cut-off) in LipidMaps.
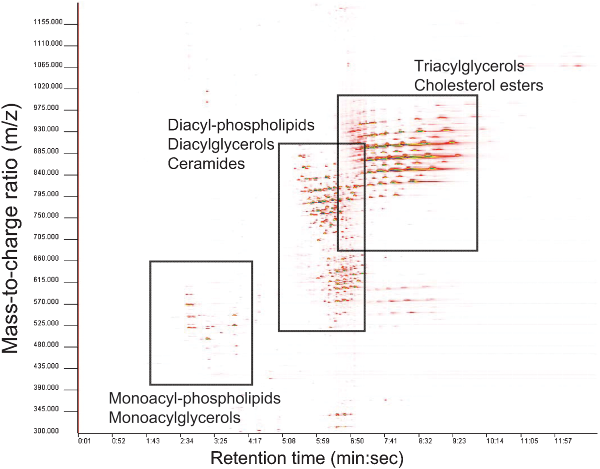
- LC-MS including HPLC-Chip/MS System, UPLC/MS, UPLC/FT-MS, LC-TOF/MS can be used to generate high resolution lipid profiles for lipids in the mass range 100-2000 Dalton. Using chromatography allows a better separation from plasma or tissue interferences, but also requires a longer run-time and handling of solvent systems.
Picture (CC-by) thanks to : Pietiläinen KH, Sysi-Aho M, Rissanen A, Seppänen-Laakso T, Yki-Järvinen H, Kaprio J, Oresic M. in Acquired Obesity Is Associated with Changes in the Serum Lipidomic Profile Independent of Genetic Effects – A Monozygotic Twin Study;PLoS ONE 2(2): e218. doi:10.1371/journal.pone.0000218
-
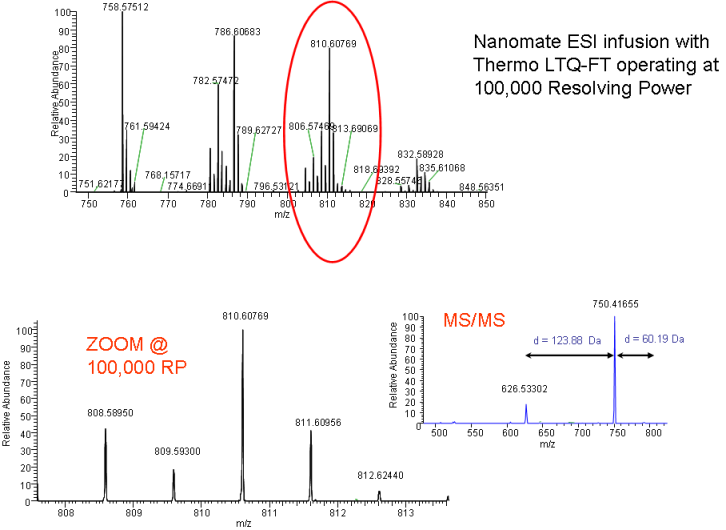
nano ESI-FTMS (chip based nano ESI FT-MS infusion) can be used for lipid fingerprinting, rapid classification, semi-quantification and structure elucidation of lipids. Using NanoMate chip based infusion together with high resolution FT-MS or Orbitrap technologies allows quick profiling (1 minute acquisition time) of lipids from crude extracts of plasma or tissue samples (plants, humans, bacteria, algea).
Picture (CC-by) thanks to: Seema Datta (Acquisition) and Tobias Kind (Evaluation) obtained on a Thermo Electron LTQ-FT-Ultra with NanoMate ESI infusion using data dependent scanning
Databases:
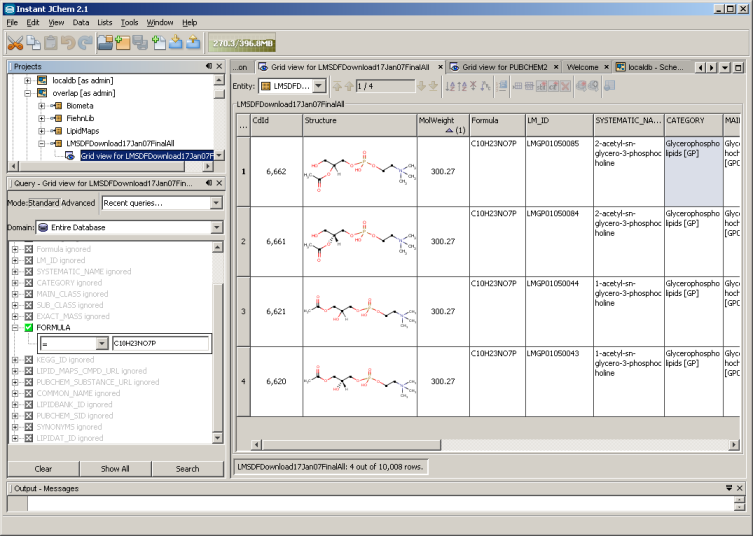
The largest curated lipid database LipidMaps contains a structure database with around 8000 unique structures (in SDF format which can be be viewed with the free Instant-JChem) which not only consist of the common free fatty acids, cholesterols, triglycerides and phospholipids, but 81 general lipid classes and 276 sub-categories. The LipidMaps consortium is based on a $35 million grant from the National Institute of General Medical Sciences (NIGMS) and supports more than 30 researchers at 18 universities. Recently researchers in Finland generated a lipid database (LipidDB) of roughly one million lipids using a scaffold library approach. Using a enumeration approach the LipidMonsterDB (lipidmaps.org) is even larger covering several million lipids.
- LipidMaps - Largest curated lipid database, containig mass spectra, classifications, protocols of 10,000 lipid species
- LipidBank - a Japanese project containing 6000 lipid species, partly with experimental information
- CyberLipids - collection of links and infos
- SphingoMap - pathway map for sphingolipid biosynthesis
- LMSAD - Lipid Mass Spectrum Analysis Database (database of sphingolipid mass spectra in XML+viewer)
- Human Metabolome DB - can be used to query for lipid species (taxonomy class lipids)
- LipidMonsterDB - is a virtual database consisting of several million lipids which can be created with lipidmapstools (thanks to Manish Sud and Eoin Fahy).
- OxPLDB - database of oxidized lipids including PAPC, POVPC, KOdiAPC, PEIPC,,PECPC, IsoketlPC, HOOAPC
- LipidHome - database of lipidomics knowledge
- LipidLibrary (AOCS) - a well curated, constantly updated and informative lipid website
Picture (CC-by): LipidMaps database from SDF file viewed on local system using the free ChemAxon Instant-JChem database. Molecular formulas, structures, names, retention indices, neutral loss fragments, accurate masses of adducts from more than 10,000 lipid species can be searched locally on a PC using one single program (MAC, LINUX, WIN).
Chemical Standards:
Chemical standards of lipids are needed for quantification purposes and as internal standards. Synthetic lipids can be used as marker substances because they are not existing in living organisms.
- www.larodan.se- pure chemical lipid standards
- www.cayman.com- pure chemical lipid standards
- www.lipomics.com - now Tethys Bioscience
- www.avantilipids.com - Avanti Polar - some with Lipidmaps ID
- www.echelon-inc.com - pure chemical lipid standards
- www.sial.com - special collection on lipids in cell signalling (Sphingolipids, Phospholipids, Inositol Phosphates, Diacylglycerols)
- www.matreya.com - sphingolipids, FAMES, glycerolipids, glycerophospholipids, tocopherols
- www.funakoshi.com - Phospholipid Kit, Natural Phospholipids, Natural Sphingolipids [PDF]
Programs and tools:
Programs for qualifying and quantifying lipids became important with the recent development of LipidOmics and the requirement to analyze the lipidome (hence all lipids). Those programs can help during the target driven setup of precursor ion scans, neutral loss scans, data dependent scans for mass spectral acquisition. Furthermore they contain features to utilize the list of 10,000 known lipids and perform data processing steps including extraction of MS/MS scans, clustering of lipids with similar MS/MS pattern and integration of monitored peaks. Please see also the other tools for comprehensive mass spectral analysis.
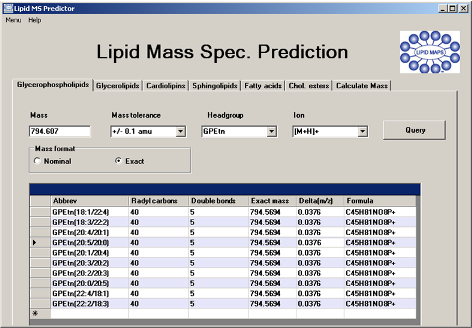
Lipid MS Predictor v1.5 (Eoin Fahy)
- LipidMaps MS Tools - calculation of m/z values, formulas, sn1 and sn2 side chains and sn3 head groups
- LipidInspector - software for lipid analysis (free beta-version available) (see DOI)
- LipidQA (Washington University Biomedical Mass Spectrometry) [Download]
- LipidProfiler LipidView (MDS Sciex)- the Lipid Profiler software uses data from the QqTOF-MS or QTRAP-MS
- LIMSA - free software for lipid analysis from University of Helsinki (see DOI)
- FAAT - software for lipid analysis using FT-MS free download here
- LipidSearch - Department of Metabolome, Graduate School of Medicine, the University of Tokyo and MKI
- Thermo LipidSearch - lipid analysis for Thermo Fisher Orbitrap mass spectrometers
- Online Lipid Calculator - Department of Pharmacology, University of Colorado Denver
- ALEX - Analysis of Lipid EXperiments (ALEX)
- TriglyAPCI - tool for interpreting atmospheric pressure chemical ionization (APCI) mass spectra of triacylglycerols (TAGs) from the Institute of Organic Chemistry and Biochemistry, Prague, Czech Republic
- LipidXplorer - for Data Dependent Acquisition (DDA) using the Molecular Fragmentation Query Language (MFQL) in shotgun lipidomics (mpi-cbg Dresden)
- AMDMS-SL (Automated Lipid Identification and Quantification by Multi-Dimensional Mass Spectrometry based Shotgun Lipidomics) (free download)
- LipidAT - Lipid Analytical Tool: automated analysis of lipidomic mass spectrometry
- STALA - Structural Analysis of Lipid A, Master Thesis Ying Ting [PDF]
- lipID - a EXCEL Add-In for phospholipid and carbohydrate analysis using FT-MS and LC-MS from FZ Borstel [PDF]
- LipidView - a software for direct infusion experiments with triple-quadrupole-instruments (QQQ)
the software is available from ABSciex.com a subsidiary of Danaher Corporation (free trial) - SimLipid - a software for identification of lipids from MS1 data, reads .txt, .xls, .mzData and mzXML.
- LipidomeDB - a data calculation environment for direct infusion- electrospray- triple quadrupole experiments
- MetISIS - a software to annotate tandem mass spectra with lipid information
- MycoMass and MycoMap - database for lipid analysis from Mycobacterium tuberculosis
- LipidMiner - a software for annotation of lipids [Sourceforge]
- MS-Lamp - a Lipidome analyzer for halogenated and sulfated lipids, the software also talks
- LipidSearch - a software tailored for Thermo instruments (Orbitrap, QQQ) (Ryo Taguchi)
- LipidLama and Greazy - software tools for automated lipid analysis
- LipidMatch - for high resolution tandem mass spectrometry experiments [PDF]
- LipidPioneer - Exact Mass Template for Lipidomics [PDF]
- LipidBlast - freely available MS/MS database for lipid identification best used as stand-alone or with MS-Dial for LC-MS/MS
- Liquid - lipid analysis tool from PNNL for HCD and CID data [software]
- LipoStar - platform independent tool with statistics, identification and pathway mappings [PDF]
- LipidHunter - tool for LC-MS/MS shotgun lipidomics assignment [PDF]
- LipiDex - software for LC-MS/MS lipid identification using MZMINE or Compound Discoverer
- LDA2 - lipid quantification and identification for multiple vendor instrumentation [PDF]
- LPPtiger - software for analysis of oxidized phospholipids [PDF]
Literature and methods:
- Lipid Analysis Methods - Book Chapters [PDF, 230 pages] [Methods in Enzymology Elsevier]
- Lipid Analysis with Andrej Shevchenko - Advion seminar
(Lipid Analysis by Nanospray Chip-Based Mass Spectrometry; Andrej Shevchenko MPI-CBG Dresden - LipidLibrary - contains a comprehensive set of literature and reviews, analytical techniques, including mass spectra, NMR techniques, Ag+ chromatography
- Multiplexed Lipid Arrays of Anti-Immunoglobulin M-Induced Changes in the Glycerophospholipid Composition of WEHI-231 Cells [PDF] by Stephen B. Milne, Jeffrey S. Forrester, Pavlina T. Ivanova, Michelle D. Armstrong, and H. Alex Brown the independent XLS sheets can be obtained from http://www.signaling-gateway.org/data/lipid
- Molecular characterization of the lipidome by mass spectrometry; Christer Stenby Ejsing;
Dissertation 2007;Technische Universität Dresden [PDF] - Schistosomal lysophosphatidylserine: an immunomodulatory factor; Kim Retra
Proefschrift 2007; Utrecht University; ISBN 978-90-393-4571-9; [PDF] [RCM] - Characterization of Molecular Glycerophospholipids by Quadrupole Time-of-Flight Mass Spectrometry: Kim Ekroos; Dissertation TU Dresden 2003; [PDF]
- An automated workflow for rapid alignment and identification of lipid biomarkers obtained from chip-based direct infusion nanoelectrospray tandem mass spectrometry (ASMS 2008) [PDF]
- Algal and bacterial lipids - Research Seminar Tobias Kind [PPT]
Additional data sources (provided for your convenience):
- LipidMaps DB 2008 as information enhanced EXCEL file (19 MB ZIP) [XLS] [small XLS]
- List of 41,510 Glycerophospholipids (GPA, GPCho, GPEtn, GPGro, GPIns, GPInsP, GPSer) in
an information enhanced EXCEL file (27 MB ZIP) [XLS] - Oxy-lipids under investigation at WHNRC John Newman's group (DiHOME, DiHET, HEPE, HETE, HODE, HOTE, EpODE, EpOME, oxo-ETE, PGF1a, PGE1, PGE2, PGE3) [DOC]
- List of accurate masses from fatty acids for mass spectrometric analysis (MS and MS/MS)
from C1 to C50 and double bonds n1 to n9 and general fatty acyls and alkyls [XLS] - A list of 444,080 monoacylglycerols, diacylglycerols and mostly triacylglycerols in a file called glycero-lipids-monster-db (168 MByte) can be found on the MAG-DAG-TAG site
- List of lipids compounds in NIST Blood Plasma SRM 1950 with absolute values as [XLS]