MS/MS or MSn fragmentation data contain a tremendous variety of information. But this information is hidden behind a curtain of rearrangements and fragmentations and ion physics and gas phase chemistry reactions we do not yet understand. Mass spectral library searches using mass spectra and MS/MS fragmentation patterns from known small molecules are routinely performed in every mass spectrometry lab. There are also some common misunderstandings regarding small molecule science and metabolomics which need to be clarified.
A) Small molecules (metabolomics, < 2000 Da) can not be sequenced like peptides or proteins (proteomics) because they do not only consist of a set of amino acids (and modifications). The good performance of de novo peptide sequencing strategies via tandem mass spectrometry does not refer to the millions of small molecules. However, small molecules consist of a very large set of diverse scaffolds. Even knowing all the scaffolds will not solve the problem in the first place. Saying that MS/MS fragmentation data can currently solve all our problems is wrong. It may be helpful in the near future. There exists currently no software for small molecules (excluding peptides) which comes near the success of de-novo sequencing programs.
B) We are not talking about drug metabolite studies and degradation products or metabolism studies, where MS/MS data or data dependent scanning or neutral loss data can identify important fragments which can supply information about hydroxylation, glucuronidation or demethylation state of known drugs. There exists software which can help with the identification of conjugates and a small subset of such metabolite transformations is known. These approaches can not help during the identification of millions of unknown and known compounds which are collected in large molecule databases like PubChem.
C) We are not talking about a known molecular structure and later try to explain the fragmentations with our mass spectral knowledge. We talk about millions of unknown substances from complex matrices like plant extracts, blood or tissue. Showing that we can detect some specific fragmentation patterns does not mean we can utilize MS/MS fragmentations for the detection of millions of unknowns. We currently can use MS/MS fragmentation data for structure elucidation of some molecules. In fact more than 99% of common metabolites in complex matrices can't be annotated or are still unknown.
MS/MS data can be currently utilized very well for the identification of unknowns in carbohydrate research and lipid research. There are approaches under development in our laboratories which will utilize mass spectral fragmentation data for fast and comprehensive automated structure elucidation of all small molecules. Such complex experiments can be performed with the LTQ-FT-Ultra and other instruments.
- MSn scan mode or MS experiments, including selected ion monitoring (SIM), selected reaction monitoring (SRM), multi-stage full scan experiment or a consecutive reaction monitoring (CRM) experiment.
- Data Dependent MS/MS experiments which select one or more ions of interest to perform subsequent scans on, such as MS/MS or ZoomScan.
- Nth Order Triple Play experiment which includes a independent scan followed by a MS or ZoomScan on the Nth most intense ions and followed by MS/MS or fullscan or dependent scan on scan event one.
- Data Dependent neutral loss MS3 experiments where a MS3 full scan is performed if a specified neutral loss is seen in the MS/MS scan.
- Data Dependent Zoom Map experiments
- Total Ion Map experiments, which can include Total Ion Maps, Neutral Loss Ion Maps, Precursor Ion Maps (which maps all precursor ions that produce the desired product ions)
- Data Dependent Ion Tree experiments, which perform data dependent MSn scans on up to 25 ions per scan over a whole scan range.
Practical example: An ion map experiment from cobalamin; on each m/z value over the full scan range a MS/MS experiment is performed (out of some thousand scans only three shown) and another experiment with a data dependent triple play with MS2 and MS3 precursor data.
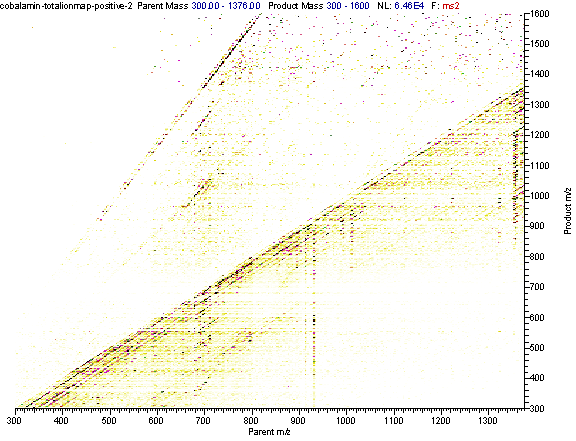
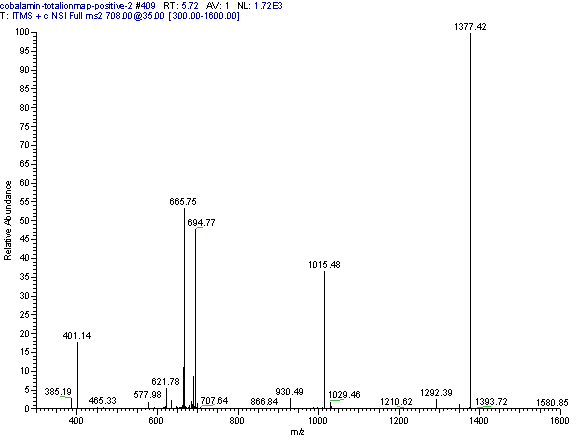
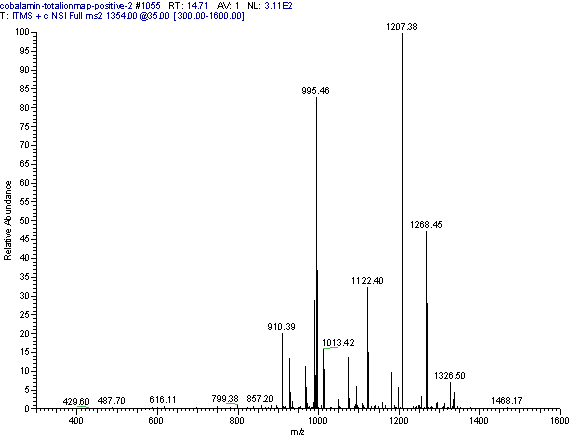
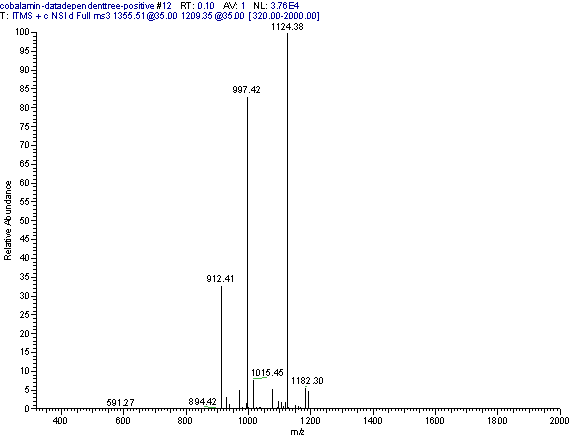
Additional links: (please see my Structure Elucidation Page for tools)
- Automated assignment of high-resolution collisionally activated dissociation mass spectra using a systematic bond disconnection approach
- Towards a full reference library of MSn spectra. Testing of a library containing 3126 MS2 spectra of 1743 compounds
- Determination of molecular structures using tandem mass spectrometry
- Maximizing MS/MS Fragmentation in the Ion Trap Using CID Voltage Ramping
- An Automated Method for Verifying Structure-Spectral Consistency Based on Ion Thermochemistry
- Ab Initio Prediction of Molecular Fragments from Tandem Mass Spectrometry Data
- Molecular Formula Analysis by an MS/MS/MS Technique To Expedite Dereplication of Natural Products
- List of Biotransformations in drug research and Common Biotransformation m/z Changes Following Enzymatic Reactions
- Detection and Characterization of Metabolites in Biological Matrices Using Mass Defect Filtering of Liquid Chromatography/High Resolution Mass Spectrometry Data
- The use of MS classifiers and structure generation to assist in the identification of unknowns in effect-directed analysis
- Mass Spectral Metabonomics beyond Elemental Formula: Chemical Database Querying by Matching Experimental with Computational Fragmentation Spectra
- Automated molecular formula determination by tandem mass spectrometry (MS/MS)
- X-Rank: a robust algorithm for small molecule identification using tandem mass spectrometry
- In silico fragmentation for computer assisted identification of metabolite mass spectra
- Identification of metabolites by using fragmentation library ("Ojimatrix") in MassBank