This theoretical example from the subset of natural compounds shows that the Seven Golden Rules are not only useful for some selected experimental examples. Around 1200 elemental compositions in the mass range from 92-2000 u were randomly selected from the KEGGdatabase, the Dictionary of Natural Compounds, Lipidmaps and many other databases. The random selection ensures that the molecular weight distribution is kept stable over the whole range.
Errors were simulated for ±3 ppm mass accuracy and ±5% isotope abundance error assuming a 3-sigma error ; hence 99.73% of all values are in the error range. Such values can be easily reached with TOF-MS.
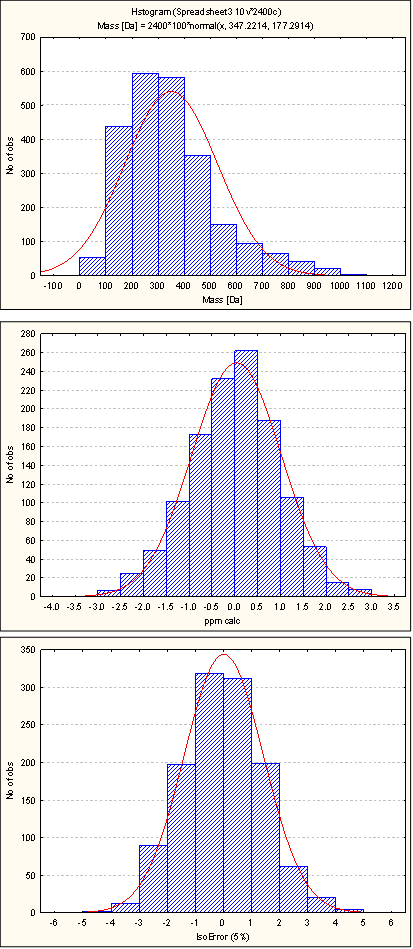
Natural Product TestSet
| Number of formulae | 1200 |
| Mass range | 92-2020 u |
| Error mass accuracy | ±3 ppm; |
| Error isotope abundances | ±5% |
| Correct identified with target DB | 99% |
| Correct identified in PubChem | 84% |
| False positive in PubChem | 10% |
| Correct identified in top three | 81% |
| Total time formula generation HR2 | 540 seconds |
| Average time for HR2 | 0.45 seconds |
| Time overhead by EXCEL | ~12h |
| Total formulae calculated | 22,966,898,012 |
| Total formulae kept by 7GR | 383,768.00 |
| Formulae reduction | -99.998% |
Download the project data as [ZIP].
An additional experiment assuming a even higher error of ±10 ppm and still ±5% isotopic abundances resulted in: 67% of the formulas were correctly annotated in PubChem, 64% correctly found in global top 3 hits and 94% correctly annotated in the natural compound target database (you need minimum version number v46 if you want to repeat the results.)
One interesting artifact revealed that the current implementation of the 7GR has problems with compounds which have 6 or more chlorines and bromines. Usually the computational time for elemental compositions up to 2000u is between 0-3 seconds, seldom exceeding 50 seconds. However for the formula C34H16Br6O9 which refers to Prunolide P (no CID) the brute force formula calculator HR2 calculated 447543 elemental compositions in 1006 seconds by evaluating 49,225,573,875 formulae (49 billion). Here the 3 isotopic abundances are obviously not enough. Also the highest peak in the mass spectrum would be at 1047.58944 u and not at 1041.5933 u (which is the isotopic mass) due to the mass defect.
Another interesting artifact was C82H86Cl4N8O29; 1786.42545 u; Kibdelin B, Ristomycin A glycone derivate, disregarding the 7GR the brute force formula calculator HR2 evaluated (in a delirium?) more than 2.077E+12 (2 trillion) formulae in 12 hours and kept 16,656,929 formulae. The entered element ranges were C:0-150 H:0-320 N:0-32 O:0-64 F:0-12 P:0-12 S:0-12 Cl:0-14 Br:0-10 and a mass range of 10 ppm. However astonishing besides the sheer number of formulae was the 124,698-fold reduction to 16,656,929 formulae among them the correct solution; evaluated externally and found by target DB match.
Here solutions which take all isotopic peaks into account and have a more sophisticated solution towards halogen compounds are clearly in advance. However be advised that there is currently no universal reverse-approach for calculating the correct elemental compositions directly out of the isotopic pattern.
Additional Link:
Chemie, Biologie und medizinische Anwendungen der Glycopeptid-Antibiotika