A Molecular Formula Generator generates possible elemental compositions from an given molecular mass. Usually a certain mass range (in ppm, in Da) must be entered to generate formulae in this range; also elements which should be included and excluded must be named. The main problem with all formula generators is: they produce too many wrong formula candidates and some of the generators are very slow. Hence chemometric rules must be applied to constrict the number of molecular formulas.
The Seven Golden Rules use heuristic rules for limiting the number of formulas only to the most probable ones. As these rules must be universal (no cherry picking or magic “pre-knowledge” allowed) they were developed and confirmed using a large dataset of 750,000 formulae covering more than 10 million known and existing compounds (PubChem, Wiley, NIST, DNP, KEGG, Beilstein, CAS). The free molecular formula generator HR2 which is used for the Seven Golden Rules is a speed-enhanced version of HiRes MS (developed by Joerg Hau) can be downloaded here.
Seven Golden Rules:
- restrictions for the number of elements,
- LEWIS and SENIOR chemical rules,
- isotopic patterns,
- hydrogen/carbon ratios,
- element ratio of nitrogen, oxygen, phosphor, and sulphur versus carbon,
- element ratio probabilities
- presence of trimethylsilylated compounds.
The Seven Golden Rules use routines from MWTWIN and HiRes for calculating some these properties. Especially the further development of the brute-force formula calculator HR2 allows counting molecular formulas in certain mass range with very high speed (usually 10 million formulas per second on an Intel Core Duo 2.0 GHz or AMD Opteron 2.8 GHz). The Seven Golden Rules are currently limited to elements C, H, N, S, O, P, F, Cl, Br, Si (the most common elements) and also exclude salts and odd electron compounds (like nitroso compounds).
Example output from HR2 (50 billion formulae evaluated - speed around 50 million formulas per second).
1073 formulas found in 1 seconds by evaluating 82,826,316 formulae.
RDBs are overloaded to maximum valence values (N=3,P=5,S=6).
19,308 formulas found in 1028 seconds by evaluating 50,533,733,862 formulae.
RDBs are overloaded to maximum valence values (N=3,P=5,S=6).
Download the free Seven Golden Rules software for elemental composition determination of small molecules here [LINK].
There are many free and commercial formula generators. One of the best free formula generators is the
Formula Finder included in MWTWIN. The problem with all formula finders is, that the number of formulae explodes in higher mass ranges (above 500 u) and sometimes several hundred thousand formulae are generated for a single accurate mass.
The following output is provided by MWTWIN (first 3 columns only), given a mass of 234.0 Da and 1 ppm window and
allowing the elements CHNSOP.Compounds found: 5
| Formula | accurate mass (u) | mass error (ppm) | LEWIS/Senior OK |
element ratio checks OK | Formula EXIST |
| CH7N4O8P | MW=234.0001512 | dm=0.6 ppm | YES | NO | NO |
| C5H4N3O8 | MW=233.9998404 | dm=-0.7 ppm | NO | NO | NO |
| C11H8O2P2 | MW=233.9999528 | dm=-0.2 ppm | YES | YES | YES |
| C12H2N4S | MW=234.0000172 | dm=0.1 ppm | YES | NO | NO |
| HN11O3P | MW=234.0001466 | dm=0.6 ppm | NO | NO | NO |
Among these compounds are 2 formulas which are not valid, because no set of structural isomers can be generated from them. Such a test can be performed with mathematical rules (LEWIS and SENIOR) or the formulas can be entered into structure generators or molecular isomer generators like MOLGEN, CDK, SMOG. The other 2 formulas are not valid, because certain high probability element ratios are not given. The only existing molecular formula (with a high probability) which can be used to construct constitutional isomers is C11H8O2P2.
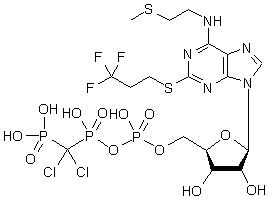
Cangrelor
C17H25Cl2F3N5O12P3S2
MW = 774.94831
A good test for the general performance of a formula generator is the drug Cangrelor (C17H25Cl2F3N5O12P3S2). The isotopic mass is 774.94831 u with elements CHNSOPFClBr. Now test your prefered formula generator without setting manual ranges for element counts or so called
*cherry picking* with a ±1 ppm or ±5 ppm error. Depending on the program, they will either never finish the calculation or they will abort after 200 or 20,000 solutions. Infact more than 44,000 formula solutions can be generated within a ±5 ppm error. The problem now is to select the right candidate out of these 44,000 possible elemental compositions. The
Seven Golden Rules can tackle this problem by limiting the element search range and matching all formulae against an internal target database (which contains known structures).
There is a strong argument for an additional orthogonal filter like
accurate isotopic abundances; used since more than 50 years in mass spectrometry and confirmed by mathematical and chemical rules in our
BMC Bioinformatics paper "Metabolomic database annotations via query of elemental compositions: Mass accuracy is insufficient even at less than 1 ppm" - check the
supplement pages for more information. Other ways would be to include mass spectral fragmentation "knowledge" or other physico-chemical constraints or chemical and mathematical rules.
Additional links:
Molecules in Silico: The Generation of Structural Formulae and Its Applications
Fully Unsupervised Automatic Assignment and Annotation of Sum Formulae for Product Ion Peaks, Neutral Losses in MS and Product Ion Spectra