The automated m/z-lookup function in LipidBlast is the quick-and-dirty approach for lipid assignments. Generally spoken, one can use just the accurate masses from a chromatographic run and assign the associated general lipid species. Conclusions about the true molecular structures of lipids such as sn1, sn2, sn3, sn4 side chains can not be made. Also depending on the resolving power multiple isobaric overlaps may occur. The approach can change to quick-and-accurate if retention information is added. Using the retention time as an additional orthogonal filter removes many of the false positive assignments. Such an approach is recommended for TOF or Exactive type instruments that just provide MS1 information. It is recommended to use accurate mass and high-resolving power instruments for the simple m/z lookup. Warning: The m/z lookup function does not utilize the awesome power and specificity of product ion information.
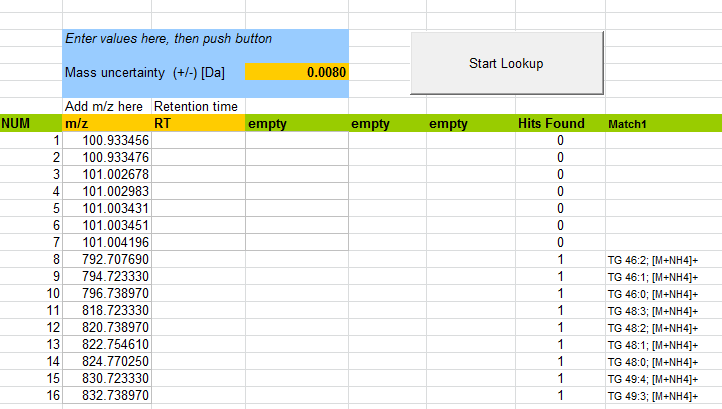
The m/z-lookup sheets in the file LIPIDBLAST-mz-lookup-v49.xls are separated into positive and negative mode. The m/z precursor information has to be extracted from the raw files or from the MGF files. The accurate masses can be extracted using Textpad or NotePad++ by marking the "pepmass" line and using the "copy bookmarked lines" functions. after the m/z values are inserted into the datasheet the experimental mass accuracy of the instrument has to be added. For well calibrated TOF or Orbitrap instruments 0.005 Da is a reachable value, for other instruments this has to be determined by external reference compounds or known expected lipid species.

The figure above shows bookmarked lines (blue triangle) that can be exclusively copied using TextPad or NotePad++. The EXCEL macro then compares all accurate mass values against the internal database which contains lipid class, carbon number and double bind count and the associated adduct information. Hits on the same row are all equivalent because all species in a certain m/z window are given back. No scoring via product ions and fragments is performed.
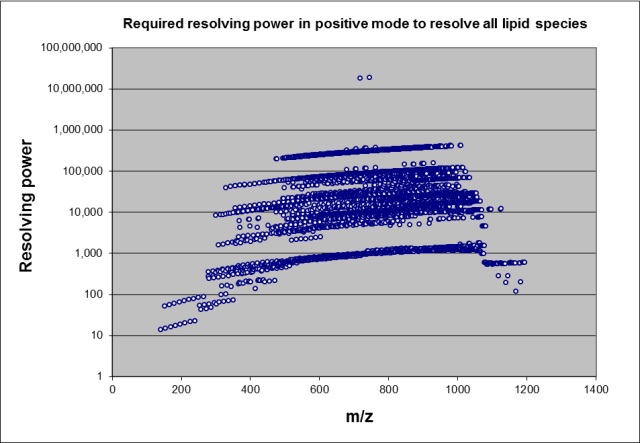
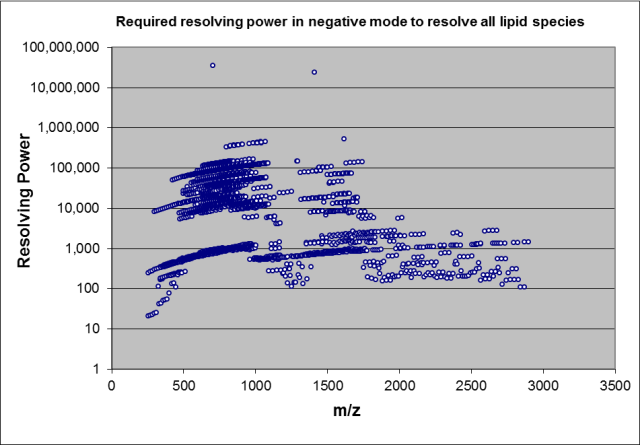
The above figure shows the required resolving power for lipids in LipidBlast using MS1 data only (complete peak resolution required). For example an instrument with 400,000 resolving power could annotated 90% of all five thousand unique isobaric masses. Ten percent of the compounds require infinite resolving power due to their isobaric masses. The problem of isobars can be prevented with using a less diverse compound space. Odd carbon chain lipids are less abundant in human plasma, but highly abundant in bacterial lipids. Excluding such compounds is easy with the included m/z-lookup database. If no Lithium modifier us used, M+Li adducts need to be excluded. The EXCEL sheets pos-mz and neg-mz contain AutoFilters that can be simply filled down by the end-user. Enable Autofilter [M+Li]+ (hence only show [M+Li]+) and select in the column USE all values with "N". Similar for plant galactolipids such as MGDG, DGDG and SQDG, exclude those if researching mammalian lipids, include them in case of algal or plant lipids. The compound classes have to be selected carefully. For example cardiolipins (abundant in animal heart tissue) are also found in plants.
Links and downloads: