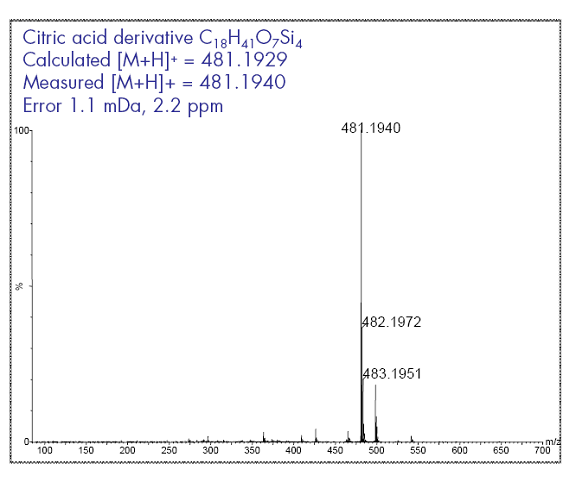
This example shows the utility of accurate isotopic abundances and accurate mass from a GC-TOF (Waters GCT Premier). In metabolomic analysis polar molecules are often derivatized by MSTFA. This reagent protects all the hydroxy groups and amino groups with a TMS (trimethyl silyl group). If a molecule has several polar moieties different silylation states can be obtained. Here a soft ionization technique helps to detect a molecular ion (which is not seen under electron impact mass spectrometry) and furthermore the correct elemental compositions can be calculated.
Seven Golden Rules
The Seven Golden Rules detected 62 molecular formulae (from elements CHNSOP) within a ±5% isotopic abundance error and ±5 ppm mass accuracy. Only one result was found in the internal elemental composition database of existing molecular formulas. This formula C18H40O7Si4 directly refers to 15 hits in the CSLS lookup service. Among them is the correct molecule of 4TMS-citric-acid.
The problem by directly searching the TMS formula C18H40O7Si4 is that this formula can only be found in specialized mass spectral databases. Subtracting the TMS groups and directly search the original compound formula of citric acid C6H8O7 would allow a constraint search in natural product databases like DNP or KEGG. Hence by getting the correct number of silicon atoms one can subtract the TMS group contribution C3H9Si and forming the original non-silylated molecule. These original formulas can then be searched in the 7GR or in CSLS.
The 7GR would calculate 60 clean formula candidates out of the 62 possible TMS hits.
C15H32N10OSi4 - (4x C3H9Si) = not possible (not enough H atoms)
C18H40O7Si4 – (4x C3H9Si) = C6H8O7 (citric acid - correct)
C13H37N8O2PSi4 – (4x C3H9Si) = CH5N8O2P (unreasonable, not allowed by 7GR)

By performing a query of all these cleaned formulae against the internal natural compound formulae only one hit will be obtained which points to possible isomer structures, among them the correct candidate citric acid. By using the “Controller page” of the 7GR software the isotopic pattern error and patterns itself must be set to 100 to allow just a internal database check. The score function (here showing a score of -191) has no meaning during this operation.
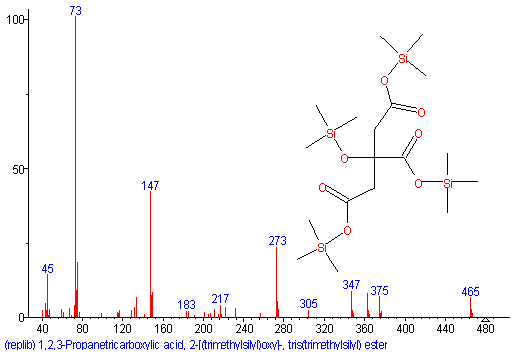
The first picture of chemical ionization on a Waters GCT Premier, CI positive (ammonia reagent gas) shows only one major peak. For comparison an electron impact (EI) ionization shows more fragmentation, good for library search and structural identification processes, however lacks the molecular ion, hence no correct elemental composition can be calculated. A GC-MS with accurate isotopic abundances and accurate mass and resolving power > 5000 together with an option for EI and CI (and MS/MS option) is the candidate of choice for GC-MS based structure elucidation.
Specification:
Citric Acid 4-TMS; PubChem CID: 553132,
Additional Links:EXACT MOLECULAR MASS DETERMINATION OF POLAR PLANT METABOLITES USING GCT WITH CHEMICAL IONIZATION
InChI: (insert as Textfield)