Project:
How large is the metabolome? A Critical Analysis of Data Exchange Practices in Chemistry
Project Partner:
Tobias Kind, Martin Scholz,
Oliver Fiehn

Picture (public domain): US long grain rice (USDA/Weller)
Results:
Kind T, Scholz M, Fiehn O (2009) How Large Is the Metabolome? A Critical Analysis of Data Exchange Practices in Chemistry. PLoS ONE 4(5): e5440.; dx.doi.org/10.1371/journal.pone.0005440; Download article here [DOI] [PDF]Short Introduction:
Calculating the metabolome size of species by genome-guided reconstruction of metabolic pathways misses all products from orphan genes and from enzymes lacking annotated genes. Hence, metabolomes need to be determined experimentally. Annotations by mass spectrometry would greatly benefit if peer-reviewed public databases could be queried to compile target lists of structures that already have been reported for a given species. We detail current obstacles to compile such a knowledge base of metabolites.As an example, results are presented for rice. Two rice (oryza sativa) subspecies have been fully sequenced, oryza japonicaoryza indica. Several major small molecule databases were compared for listing known rice metabolites comprising PubChem, Chemical Abstracts, Beilstein, Patent databases, Dictionary of Natural Products, SetupX/BinBase, KNApSAcK DB, and finally those databases which were obtained by computational approaches, i.e. RiceCyc, KEGG, and Reactome. More than 5,000 small molecules were retrieved when searching these databases. Unfortunately, most often, genuine rice metabolites were retrieved together with non-metabolite database entries such as pesticides. Overlaps from database compound lists were very difficult to compare because structures were either not encoded in machine-readable format or because compound identifiers were not cross-referenced between databases.We conclude that present databases are not capable of comprehensively retrieving all known metabolites. Metabolome lists are yet mostly restricted to genome-reconstructed pathways. We suggest that providers of (bio)chemical databases enrich their database identifiers to PubChem IDs and InChIKeys to enable cross-database queries. In addition, peer-reviewed journal repositories need to mandate submission of structures and spectra in machine readable format to allow automated semantic annotation of articles containing chemical structures. Such changes in publication standards and database architectures will enable researchers to compile current knowledge about the metabolome of species, which may extend to derived information such as spectral libraries, organ-specific metabolites, and cross-study comparisons.
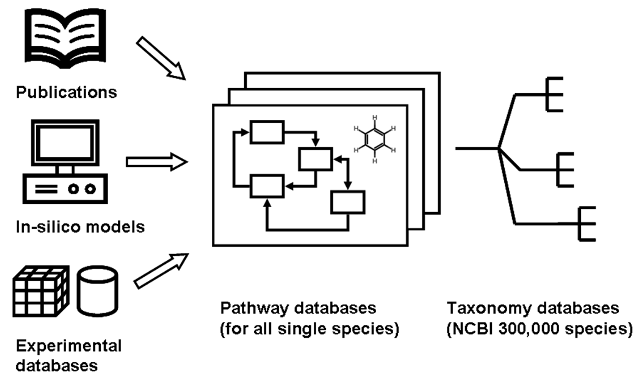
Pathway and small molecule database creation is performed from A) Publications and published papers B) in-silico data or computational models (ortholog mapping approaches) B) molecules directly obtained from experimental repositories or databases. For each molecule and pathway taxonomy data must be associated, or for each species a single database is created. Associated compartment data (organ, tissue) should be included as well as endogenous or exogenous origin of data.
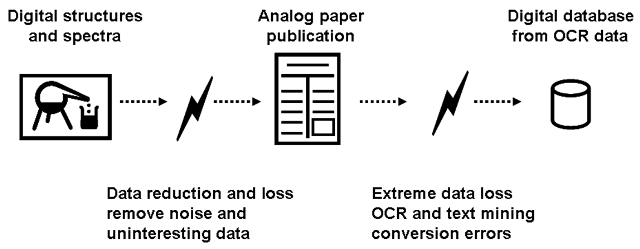
Molecular structure and spectral data is usually obtained from bitmap data using optical character and optical structure recognition. The associated data loss (hamburger-to-cow algorithm) can be avoided by direct submission of machine readable structures and machine readable spectra to large institutional repositories. OCR algorithms are valuable and needed for already published data, but a paradigm change must be initiated for new chemistry, biochemistry and metabolomics publications.
Machine readable semantic data (living cow) should not be converted into a hamburger, hence bitmap structures and bitmap spectra that are not machine readable anymore. Later algorithms are used to convert this hamburger back to a cow, which is error-prone and not needed, because original vectorized structures and high resolution spectra are available "as is" from publication authors and researchers. (Our critique is not aimed at text OCR algorithms and cloning cows from well done hamburgers may be possible in the future.) [Large Picture]
Provided project software:
Due to multiple license restrictions the data here is presented for academic research and peer-review only. By downloading, you agree to use the data either for academic research or peer-review. Download the supplement data and project software here EXCEL 2000 [XLS] or [ZIP]. The file can be opened with Microsoft EXCEL or OpenOffice.
Picture service (for your convenience):
Parts of the software supplement of the publication are published under the
Creative Commons (by) license. This license lets others distribute, remix, tweak, and build upon your work, even commercially, as long as they credit you for the original creation. Download PPT and TIFF pictures as [ZIP].Call for participation and open discussion:
Please comment on this article or discuss barriers, problems, obstacles or missing projects from the article. The focus is not on how many databases exist, but how such databases can be enriched with experimental, machine readable data from electronic structure and spectral data submissions directly from publications. The PLOS comment section requires a valid (non-anonymous) login, the comment rider is on the top see below graphics [LINK to comments].

Please comment on the article by creating a PLOS login and write about your ideas regarding this article.
Links to external software used in the project:
Programs:
- TEXTPAD ($$) www.textpad.com
- MS EXCEL ($$$) + Visual Basic www.microsoft.com
- ChemAxon molconvert (free), cxcalc (academic license), JCHEM full (academic license)
- ChemAxon Instant-JChem (free academic version)
- EPA EPISuite (free)
- Beilstein Crossfire for searching the Beilstein database of organic compounds and properties
- Scifinder Scholar for searching the CAS database
- InChI and InChIkey software (free)
Databases and Services (updated):
Compound annotations from text (Name; PubChem CID; InChIKey):
2-acetyl-1-pyrroline;
CID 522834;
DQBQWWSFRPLIAX-UHFFFAOYAG
Vitamin-A; CID
445354;
FPIPGXGPPPQFEQ-OVSJKPMPBW
Beta-carotene; CID
5280489;
OENHQHLEOONYIE-JLTXGRSLBT
Bisbynin; CID NA;
ICHJNTDKHBXTFN-CMZGOGIXBZ [CML] [MOL] [ChemSpider]
Trans-luteine; CID
5368396;
KBPHJBAIARWVSC-DKLMTRRABK
Cholesterol; CID
5997;
HVYWMOMLDIMFJA-DPAQBDIFBB
Malathion; CID
4004;
JXSJBGJIGXNWCI-UHFFFAOYAK
Chlorpyrifos; CID
2730;
SBPBAQFWLVIOKP-UHFFFAOYAG
Ribosylnicotinamide; CID
439924;
JLEBZPBDRKPWTD-ARWKKGFBBE
Omeprazol; CID
4594;
SUBDBMMJDZJVOS-UHFFFAOYAZ
Rhodopinal; CID
20055178;
GOJQFVQXKNNAAY-XQHLYSSHBM
Tegafur; CID
5386;
WFWLQNSHRPWKFK-UHFFFAOYAE
Arginine; CID
232;
ODKSFYDXXFIFQN-UHFFFAOYAT
Optical Character Recognition and Chemical Structure Recognition:
- OSRA - Optical Structure Recognition (NIH) (free, open source)
- Kekule - OCR-optical chemical (structure) recognition (NCI)
- Clide & Clide Pro - Chemical literature data extraction tool (Univ. Leeds/ SimBioSys/Keymodule)
- ChemoCR - Tool for Chemical Compound Reconstruction
- ChemReader - Automated extraction of chemical structure information
Text based semantic annotation tools and projects:
- Oscar3 - Open Source Chemistry Analysis Routines (open source)
- Chem-MANTIS - Nomenclature Transformation Integrated System
- Project Prospect - IUPAC, Ontology, CML, InChI enhanced chemical publications
- Chemicalize.org - web based annotation service via ChemAxon proxy (name to structure)
Name to chemical structure converters (vice versa):
- Autonom - Beilstein Institute
- IBM Chemical Annotator - IBM Almaden
- Lexichem - OpenEye
- Struct <=> Name - CambridgeSoft
- Marvin IUPAC Name - ChemAxon
- ACDName - Structure to Name and Name to Structure ACDLabs
- NameExpert and Nomenclator - Cheminnovation
- IUPAC NameIt - BioRad
- OPSIN - name to structure converter open source project (OSCAR3)