The number of MS/MS scans in MGF files can easily exceed 5,000 to 10,000 product ions, a manual inspection is timewise not possible. Therefore automated batch search and scoring is important for a streamlined search. The NIST MS PepSearch program is a command-line version that is related to the NIST MS Search GUI program. Originally aimed at peptide scoring it can also be used for small molecule scoring. The search speed for LipidBlast libraries is usually around 1000 spectra/second, hence even millions of MS/MS spectra can be searched in a reasonable time. One has to be aware that the search scores from the NIST MS Search GUI are different to those obtained from the NIST MS PepSearch.
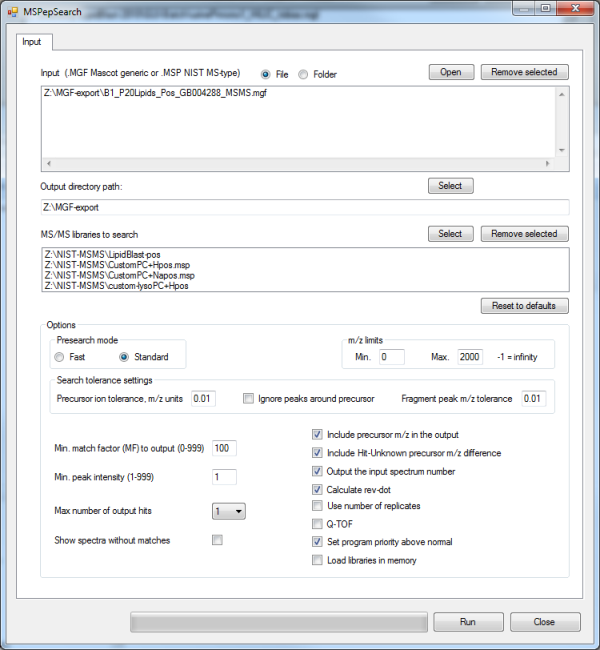
The input files are MGF files, there are 4 NIST MS/MS libraries that can be searched and parameters must be set according to the instrument definitions. The generated *.TSV file (tab separated file) can be directly imported into MS EXCEL or the free Open Office Calc program. In EXCEL the Data--> Filter --> Autofilter function can be used to select and sort reveres dot-product scores and refine additional parameters. The following picture details the XLS output.
The lower search score limits have to be defined by reference compounds measured on the specific instrument and the library selected. For LipidBlast it is recommended to use the Rev-Dot (reverse dot) scores and additionally also match compounds according to their elution order and retention time. Multiple adducts for the same compound can act as a confirmation. For example lysoPC 18:2 in the above figure is found ans [M+H]+ and [M+Na]+.
Because LipidBlast covers algal, bacterial and animal lipids, a specific taxonomy filter has to be applied. For example MGDG or DGDG or SQDG are not commonly found in human blood plasma. But these plant specific lipids are always existing in plants. Because LipidBlast also covers a large range of adducts, such adducts have to be enabled or disabled too. For example M+Li adducts are usually only observed if Lithium is directly added to the extract or solvent. If the LC-MS/MS run uses acetate buffer, the formate adducts have to be excluded.
In case of a fast scanning instrument with multiple MS/MS scans per MS1 a precursor ion exclusion times has to be set, otherwise the same precursor is fragmented repeatedly. If the same precursor mass is physically separated by a few seconds or minutes there is a high chance that different isomers were physically separated. In any case the final list should be investigated in the NIST MS Search GUI. The numbers {Num} in the first column refer to the spectral number in the MGF file. These numbers are correlated, hence a visual inspection of the found hits is easy.
The current GUI version (2014) only allows 4 MS/MS libraries at once, in order to search 10 or more libraries, use the *.TSV file as a template, open it, the second line contains the commandline parameters. Open the Windows command prompt by typing CMD in the runbox and execute MSPepSearch.exe on the command line directly. Of course all additional libraries have to be added manually by adding the /LIB parameter and the full path of the library in quotes.
LINKS and downloads
- NIST MS PepSearch [LINK]