What is better: PLS, OPLS or PLS-DA?
A recent TRAC article Notes on the practical utility of OPLS
(DOI: 10.1016/j.trac.2009.08.006) by Tapp and Kemsley discusses:
[1] S. Mahadevan, S.L. Shah, T.J. Marrie and C.M. Slupsky, Anal. Chem. 80 (2008), p. 7562. Full Text via CrossRef | View Record in Scopus | Cited By in Scopus (27) Recently, Mahadevan et al. [1] mistakenly concluded “…in the case of metabolomic data sets where there is a significant divergence in the within-class variation between the two classes, OPLS-DA might perform better than PLS-DA”. However, like-for-like comparison will show that OPLS-DA never outperforms PLS-DA, just as OPLS will never outperform PLS.
They write that OPLS has "no predictive performance advantage over traditional PLS". They discuss PLS-DA, OPLS-DA and a set of 60 references and literature analysis is given together with some mathematical explanation. I have never heard the term metabogram, but I know that SIMCA-P (Umetrics) is often used because its user friendly (Disclaimer: I use WEKA, R and Statistica). I am not sure if the authors disclosed in their article that they wrote their own (free) implementation of PLS, IFRNOPLS based on MatLab and Eigenvector routines.
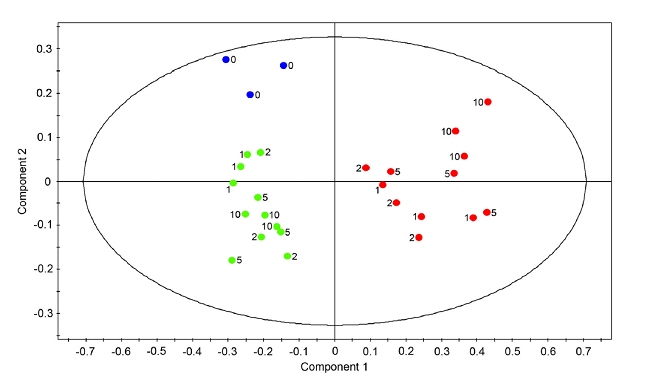
OPLS-DA example, thanks to Zulak et.al BMC Plant Biology 2008, 8:5
From a practitioners point of view I can conclude that the model choice (free or commercial) is not important as long as the algorithm is statistical sound. Furthermore, the software should allow easy data handling of metabolomics data sets with the ability of investigating the raw data, whenever possible. For supervised methods it is important to avoid overfitting by choosing a large enough validation set or with internal cross-validation. Especially PLS-DA is prone to overfitting. Other than that, I am a fan of workflows that use a series of classification models, and select the best and fastest by bagging, boosting or voting.
One final major challenge is to deploy the validated model as software for others to re-use. Here a simple classification model may rule over a complex neural network. Statistica for example has an automatic code generator for VBA and C++ (unfortunately not for JAVA) that can be directly recompiled into a program.
One final argument is the submission of raw data and the final metabolomics matrix together with the paper. In this way any misguiding claim, like OPLS is better tha PLS-DA, PLS-DA is better than PLS can be validated with a set of independent methods, like a multi-class ANOVA or a simple feature selection process (for biomarker finding) with PCA (for visualization).
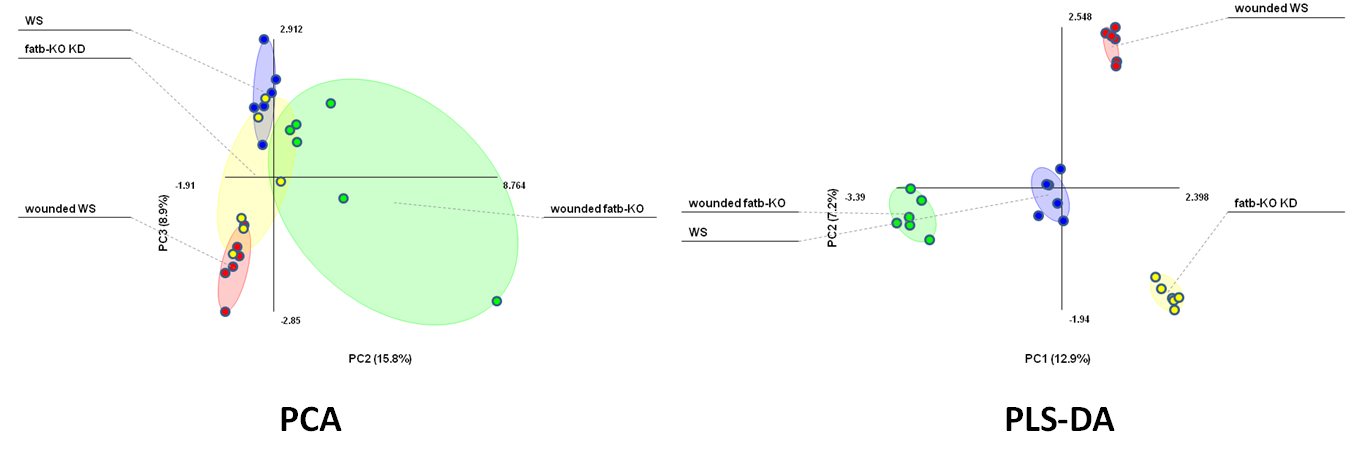
Unsupervised PCA does not separate well between classes, PLS-DA separates classes (Figure created with the free MultiBase EXCEL plugin, Data SetupX ID:115958 Fatb Induction Experiment (FatBIE) from Arabidopsis)
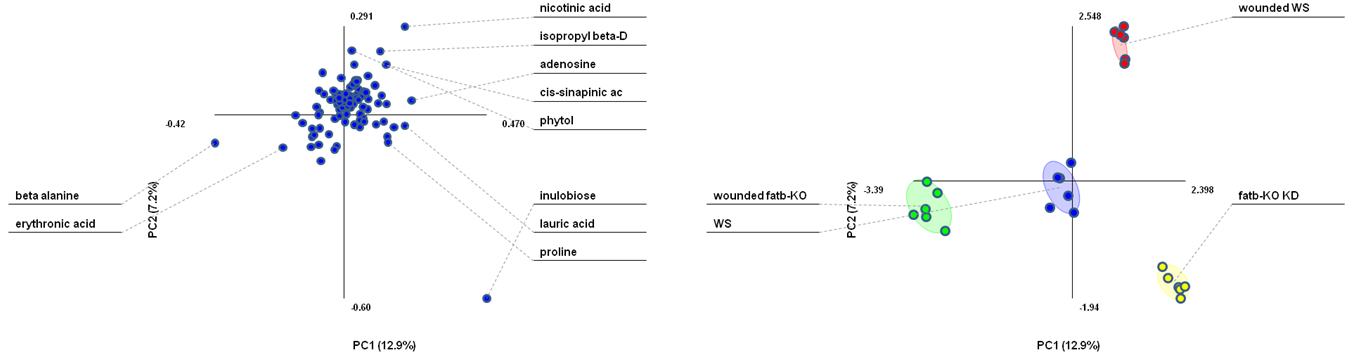
PLS-DA loadings plot (left) and PLS-DA scores plot (right). The loadings plot shows the variable influence on the separation. (Figure created with the free MultiBase EXCEL plugin, Data SetupX ID:115958 Fatb Induction Experiment (FatBIE) from Arabidopsis)
Literature and articles of interest
- Notes on the practical utility of OPLS
- Multivariate paired data analysis: multilevel PLSDA versus OPLSDA
- OPLS: an ideal tool for interpreting PLS regression models? (hosted by Eigenvector maker of the PLS-Toolbox)
- Statistical strategies for avoiding false discoveries in metabolomics and related experiments (Overfitting problem)
- Assessing the performance of statistical validation tools for megavariate metabolomics data (Overfitting problem)
Programs and Tools
- Hiroshi Tsugawa's free statistical EXCEL software for multi t-test, PCA, PLS-R and PLS-DA
- MultiBase - NumericalDynamics provides a free EXCEL plugin for PCA, PLS-DA and PLS-EDA (Download available)
- KOPLS - Kernel-based Orthogonal Projections to Latent Structures (K-OPLS) for regression and classification [PDF]
- TANAGRA - Tanagra (free) as stand-alone and with EXCEL plugin provides PLS-DA and PLS-LDA [PDF]
- MetaboAnalyst - Wishart group (Jianguo Xia) provides a free platform for PCA and PLS-DA
- IFRNOPLS - an alternative to OPLS
- Tutorial and code for OPLS (MatLab) from the BDAGroup (NL) [PDF] and [ZIP]
- SIMCA P - Umetrics provides a PLS-DA software
- Unscambler - CAMO provides a PLS-DA software
- PLS-Toolbox - Eigenvector provides a PLS and PLS-DA software
- Example data for provided graphs (SetupX ID:115958)