Project:
FiehnLib - a mass spectral and retention index library for comprehensive metabolic profiling
The current libraries comprise over 1,000 identified metabolites that are currently screened by the Fiehn laboratory. We are continually extending the compound list and welcome compound donations or compound names with PubChem CIDs that are yet missing in our lists.
If you are interested in obtaining the Fiehn mass spectral libraries, you can purchase these for GC-quadrupole mass spectrometers from Agilent, and for GC-TOF mass spectrometers from Leco. (see links below)
You can also directly compare mass spectra by compound names, by database identifiers or even against your own GC/MS spectra by querying BinBase.
Project Partner:
Tobias Kind, Mine Palazoglu, Do Yup Lee, Yun Lu, Gert Wohlgemuth, Martin Scholz, Oliver Fiehn
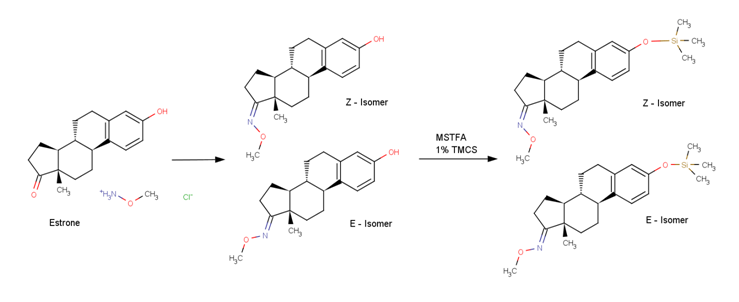
Estrone CID: 5870 is first methoximated resulting in two stereoisomers and later silylated; Two peaks are observed in the GC-MS.
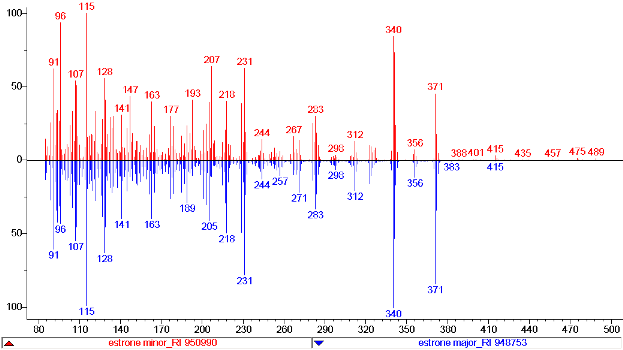
The two estrone mass spectra (Estrone major and Estrone minor) show slightly different mass spectra (m/z 207, m/z 96 and m/z 341). The different stereoisomers (E/Z or syn/anti) result in two peaks and are perfectly separated in the GC-MS. The retention time difference is almost two seconds.
Publication:
FiehnLib – mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry; Kind, Tobias; Wohlgemuth, Gert; Lee, Do Yup; Lu, Yun; Palazoglu, Mine; Shahbaz, Sevini; Fiehn, Oliver Accepted in Analytical Chemistry (ACS) - currently in ASAP section [DOI].
http://pubs.acs.org/doi/abs/10.1021/ac9019522
Provided project software:
Parts of the software supplement of the publication are published under the Creative Commons (by) license. This license lets others distribute, remix, tweak, and build upon your work, even commercially, as long as they credit you for the original creation. Download the supplement data and CC-by project software here EXCEL 2000 [XLS] or [ZIP]. The files can be opened with Microsoft EXCEL or OpenOffice.
- FiehnLib as EXCEL 2003 file with Name, PubChem CID, INChIKey,pictures [XLS]
- FiehnLib as SDF from PubChem (molecule file) [SDF]
- FiehnLib as web link for easy web access [LINK]
There are two large Open Source software projects which utilize the FiehnLib mass spectral and retention library. First the open source SetupX LIMS system which handles sample organization, class assignments and provides download web services. Secondly the BinBase automatic library search and retention index matching system which performs the identification and post-matching.
- SetupX LIMS (Open Source) - Online access - [Download Source Code]
- BinBase (Open Source) - [Download Source code]
The FiehnLib library itself with all 1200 mass spectra and retention indices and a complete procedure for metabolic profiling of tissue and plant material is licensed by different mass spectrometer producers, please follow the links below.
- Agilent Fiehn GC/MS Metabolomics RTL Library [LINK]
An Agilent 6890GC/5973 quadrupole MSD was used for the creation of the Agilent library. The GC was equipped with a DB5-MS + 10m Duragard capillary column (length 30m; ID 0.25 mm, film thickness 0.25 μm; Agilent 122-5532G). Helium in constant flow mode was used as a carrier gas. The GC temperature program was set at an initial temperature 60°C with a hold time of one minute; with a following temperature ramp of 10°C/min and a final temperature of 325°C and final hold time of 10 minutes. The MSD transfer line heater temperature was set to 290°C. The mass spectrometer was operated in full scan mode from 50 m/z to 650 m/z. Electron impact ionization at 70V was employed with an ion source temperature of 250°C. Myristic acid D27 (Sigma Aldrich 366889; PubChem SID 24862719; InChIKey: TUNFSRHWOTWDNC-RZVOLPTOSA-N) was used for retention time locking (RTL).
- LECO Fiehn Metabolomics library [LINK]
The LECO library was obtained on a Leco Pegasus IV time of flight mass spectrometer with a Agilent 6890 gas chromatograph and a Gerstel automatic liner exchange system with multipurpose sample MPS2 dual rail and two derivatization stations in conjunction with a Gerstel CIS cold injection system (Gerstel, Muehlheim, Germany). A 30 m long, 0.25 mm i.d. Rtx-5Sil MS column with 0.25 mm 95% Dimethyl 5% Diphenyl-Polysiloxane film and additional 10 m integrated guard column was used (Restek, Bellefonte PA) for separation. The GC oven temperature was held constant at 50°C for 1 min and then ramped at 20°C/min to 330°C at which it was held constant for 5 min. The transfer line temperature between gas chromatograph and mass spectrometer was set to 280°C. Electron impact ionization at 70V was employed with an ion source temperature of 250°C. Spectra were collected from m/z 85-500 at 20 spectra/sec and 1550V detector voltage. Fatty acid methyl esters (FAMEs) were used as internal retention index markers.
Links to external software used in the project:
Programs:
- TEXTPAD ($$) www.textpad.com
- MS EXCEL ($$$) + Visual Basic www.microsoft.com
- ChemAxon molconvert (free), cxcalc (academic license), JCHEM full (academic license)
- ChemAxon Instant-JChem (free academic version)
- Scifinder Scholar for searching the CAS database
- ChemAxon Marvin Sketch (free)
- NIST MS Search 2.0 (free)
- LECO ChromaTOF software (with GC-MS) ($$$$)
- Agilent Chemstation software (with GC-MS) ($$$$)
- Statistica Dataminer v8.0 (academic license) ($$$$)
Databases and Services (updated):
- The PubChem database (free) - download the whole PubChem DB here: PubChem FTP
- The Dictionary of Natural Products ($$$$) Web version
- The KEGG database (free)
- The peptide DB and metabolome DB (free)
- The MDL Beilstein database ($$$$$)
- The CAS database ($$$$$ academic or $$$$$$ commercial)
- The ChemSpider DB (free) - largest information enhanced DB with mass spectrometry API
- The SetupX - biological experiment database
- The KNApSAcK DB - Species-Metabolite Relationship Database
- The LipidMaps DB - LIPID Metabolites And Pathways Strategy
- The NCBI Taxonomy DB
- The ZINC Database - A free database for virtual screening
- The MDL ACD - Available Chemicals Directory ($$$$)
- The NIST05 Mass Spectral and Retention Index Library ($$$$)
- The GOLM Metabolomics DB This email address is being protected from spambots. You need JavaScript enabled to view it. (free download)
- The MassBank DB MassBank DB (free access)
Compound annotations from publication (Name; PubChem CID; InChIKey):
Methylhydroxylamine Hydrochloride; CID: 521874; InChIKey=XNXVOSBNFZWHBV-UHFFFAOYAC
Pyridine; CID: 1049; InChIKey=JUJWROOIHBZHMG-UHFFFAOYAY
N-Methyl-N-trimethylsilyltrifluoroacetamide; CID: 32510; MSPCIZMDDUQPGJ-UHFFFAOYAZ
Trimethylchlorosilane; CID: 6397; InChIKey=IJOOHPMOJXWVHK-UHFFFAOYAM
Chloroform; CID: 6212; HEDRZPFGACZZDS-UHFFFAOYAG
Helium; CID: 23987; SWQJXJOGLNCZEY-UHFFFAOYAJ
Estrone; CID: 5870; DNXHEGUUPJUMQT-CBZIJGRNBW
(Z)-O-methoxyamino-estrone; InChIKey=PBXZSAMJKKLMEF-DUTNZQCHBS
(E)-O-methoxyamino-estrone; InChIKey=PBXZSAMJKKLMEF-YDKWXADCBY
O-TMS-(Z)-O-methoxyamino-estrone; InChIKey=GIUGKXYPAYLRPS-AFLCNQENBJ
O-TMS-(E)-O-methoxyamino-estrone; InChIKey=GIUGKXYPAYLRPS-XKNWVNBWBA
Source file for compounds in ChemAxon MRV format. MARVIN Sketch can be downloaded for free [LINK].